Professor Sherif El-Khamisy
School of Biosciences
Professor of Molecular Medicine
Deputy Director of the Healthy Lifespan Institute


Full contact details
School of Biosciences
Firth Court
Western Bank
∫˘¬´”∞“µ
S10 2TN
- Profile
-
Career history
- 2015 - present: Director of Research and Innovation
- 2014 - present: Chair of Molecular Medicine, Krebs Institute, University of ∫˘¬´”∞“µ
- 2014 - present: Wellcome Trust Investigator, Krebs Institute, University of ∫˘¬´”∞“µ
- 2014 - present: Lister Research Fellow, Krebs Institute, University of ∫˘¬´”∞“µ
- 2013 - 2015: Wellcome Trust Group Leader, Genome Center, University of Sussex
- 2013 - 2014: Reader, Krebs Institute, University of ∫˘¬´”∞“µ
- 2008 - 2013: Wellcome Trust Fellow, Genome Center, University of Sussex
- 2007 - 2008: MRC Post-doctoral Fellow, Genome Center, University of Sussex, UK
- 2006: Post-doctoral Fellow, Dept. of Genetics, St Jude Children’s Research Hospital, USA
- 2005: Lecturer, Dept. of Biochemistry, Ain Shams University, Egypt
- 2002 - 2005: PhD in Biochemistry, University of Sussex, UK
Video animations summarising recent research discoveries
This video describes how DNA repair guards us against ALS, in particular the most common genetic cause for ALS and frontotemporal dementia caused by expansion in a gene called C9orf72. It summarises the work published in Nature Neuroscience on 17 July 2017.
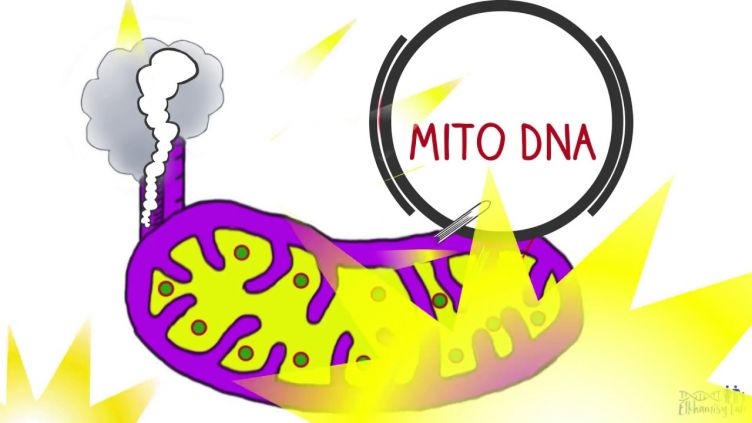
How can the cells' powerhouse - mitochondria - protect their DNA? This video summarises a recent publication in Science Advances.
Major scientific accomplishments
- Discovery of the mechanism of genomic instability and neural cell death in C9orf72 ALS (Nature Neuroscience 2017)
- Discovery of protein-linked chromosomal break repair in the mitochondria (Science Advances 2017)
- Discovery of a novel therapeutic strategy for ALS by inhibiting nuclear export (Nature communications 2017)
- Elucidation of an epigenetic mechanism underlying irinotecan resistance in colorectal cancer (Nucleic Acids Research 2016)
- Development of a nano-genomic technology to diagnose nucleic acid based infections (Biosensors 2016)
- Discovery of the first human diseases resulting from accumulation of Top2-linked DNA breaks (Nature Genetics 2014)
- Elucidation of the mechanisms underlying temozolomide therapy in brain tumors (Nucleic Acids Research 2014)
- Development of techniques to measure protein-linked chromosomal breaks (Plos One 2013)
- Identification of the molecular role of SUMOylation during single-strand break repair (Nature Communications 2012)
- Identification of the role of XRCC1 during neural development and maintenance (Nature Neuroscience 2010)
- Discovery of the enzyme that disjoins abortive topoisomerase 2 DNA breaks (Nature 2009)
- Discovery of the function of the neuroprotective enzyme aprataxin (Nature 2006)
- Identification of the first human disease with defects in chromosomal single-strand break repair (Nature 2005)
- Elucidation of the role of CK2 in chromosomal single-strand break repair (Cell 2003)
- Qualifications
-
Honours and distinctions
- Wellcome Trust Investigator Award
- Fellow of the Royal Society of Chemistry
- Fellow of the Royal Society of Biology
- Fellow of the Lister Institute of Preventative Medicine
- Shoman Award for Medical Sciences
- Sir Richard Stapley Award, UK
- Lorne Duncan Award, UK
- Research interests
-
The intertwined nature of the DNA double helix creates topological barriers that need to be resolved during all types of DNA transactions, in all forms of life from plant to man.
DNA topoisomerases achieve this by breaking and sealing the double helix, allowing the DNA to become untangled or unwound. During their normal catalytic cycle DNA topoisomerases become covalently attached to the DNA 3’-end (Top1) or to the 5’-end (Top2) via a reversible covalent phosphotyrosyl bond.
The presence of nearby oxidative DNA breaks (red circles) or collision with elongating RNA polymerases (RNA POL) during transcription results in ‘trapping’ of topoisomerases on DNA, causing protein-linked DNA breaks (PDBs), which are potent blocks to transcription and cell viability.
PDBs are repaired by a nucleolytic cleavage of DNA releasing the stalled Top and a fragment of DNA. Since this process results in an inevitable loss of genetic material, it is inherently error-prone.
Alternatively, PDBs can be repaired by an error-free mechanism in which the covalent phosphotyrosyl bond linking DNA to the stalled Top is cleaved by specific enzymes, such as tyrosyl DNA phosphodiesterases (TDP1 and TDP2). Interestingly, defects in TDPs cause neurological disease in man (see Fig.2).
In addition to maintaining genetic integrity in post-mitotic non cycling cells, accumulation of PDBs in cycling cells has been widely exploited to treat cancer (e.g. topoisomerase poisons such as Irinotecan and Topotecan), and therefore small molecule inhibitors of TDPs are emerging as attractive strategies to improve cancer therapy.
- Publications
-
Show: Featured publications All publications
Featured publications
Journal articles
- . Life Science Alliance, 5(8), e202101145-e202101145.


- . Cellular and Molecular Life Sciences, 79(3).


- . Life Science Alliance, 4(10), e202000966-e202000966.


- . Nature Communications, 12.


- . Nature Communications, 12(1).


- . Cancers, 13(10).


- . Science Advances, 7(5), eabc4165-eabc4165.


- . Oncotarget, 11(13), 1109-1130.


- . Nature Communications, 11(1).


- . Biosensors and Bioelectronics, 141, 111451-111451.


- . EBioMedicine, 35, 106-113.


- . Trends in Genetics, 34(8), 574-577.


- . Free Radical Biology and Medicine, 126, 153-165.


- . Oncotarget, 9(53), 30053-30065.


- . Cell Reports, 23(11), 3352-3365.


- . Journal of Cellular Biochemistry, 119(8), 6869-6881.


- . Brain, 141(5), 1247-1262.


- . Nature Reviews Molecular Cell Biology, 19(6), 343-344.


- . Nature Neuroscience, 20, 1225-1235.


- . Nature Communications, 8.


- . Science Advances, 3(4).


- . Nucleic Acids Research, 45(3), 1159-1176.


- . Biosensors and Bioelectronics.


- . Mechanisms of Ageing and Development.


- . Scientific Reports, 6, 26626-26626.


- . Cell Reports, 15(4), 893-908.


- . Nature Reviews Cancer, 15(3), 137-151.


- . Human Molecular Genetics, 24(3), 828-840.


- . Molecular Cancer Therapeutics, 14(2), 575-585.


- . Nature Genetics, 46(5), 516-521.


- . Nucleic Acids Research, 42(5), 3089-3103.


- . PLoS One, 8(4), e58239.


- . Nucleic Acids Res, 40(17), 8371-8380.


- . Nat Commun, 3, 733.


- . EMBO Mol Med, 3(2), 78-88.


- . J Biol Chem, 286(1), 403-409.


- . Hum Mol Genet, 19(7), 1324-1334.


- . Nature, 461(7264), 674-678.


- . Nat Neurosci, 12(8), 973-980.


- . EMBO J, 26(22), 4720-4731.


- . Nature, 443(7112), 713-716.


- . NATURE, 434(7029), 108-113.


- . Cell, 117(1), 17-28.


- . Nucleic Acids Res, 31(19), 5526-5533.


All publications
Journal articles
- . Journal of Genetic Engineering and Biotechnology, 22(4), 100427-100427.


- . Neuromuscular Disorders, 43, 104441.131-104441.131.


- . Frontiers in Cell and Developmental Biology, 12.


- . DNA Repair, 103629-103629.


- . Cancer Research, 84(1_Supplement), A023-A023.


- . iScience, 26(11), 108219-108219.


- . Nutrition Today, 58(3), 105-118.


- . Trends in Cell Biology.


- . Genes, 14(1).


- . Frontiers in Genetics, 13.


- . Nature, 609(7929), 1038-1047.


- . Seminars in Cancer Biology.


- . Cancer Research, 82(12_Supplement), 1049-1049.


- . Frontiers in Aging Neuroscience, 14.


- . Life Science Alliance, 5(8), e202101145-e202101145.


- . Cellular and Molecular Life Sciences, 79(3).


- . Pharmaceutics, 14(1).


- . Life Science Alliance, 4(10), e202000966-e202000966.


- . Nature Communications, 12.


- . Biochimie, 190, 70-90.


- . Oral Surgery, Oral Medicine, Oral Pathology and Oral Radiology, 132(1), e15-e15.


- . Nature Communications, 12(1).


- . International Journal of Molecular Sciences, 22(11).


- . Cancers, 13(10).


- . Scientific Reports, 11(1).


- . Science Advances, 7(5), eabc4165-eabc4165.


- . Stem Cell Reports, 14(6), 1009-1017.


- . Oncotarget, 11(13), 1109-1130.


- . Nature Communications, 11(1).


- . Nature Communications, 10(1).


- . Biosensors and Bioelectronics, 141, 111451-111451.


- . DNA Repair, 81, 102669-102669.


- A new filter-based Gene selection method based on dragonfly optimization and correlation-based feature selection. BIOSCIENCE RESEARCH, 16(3), 3139-3154.


- . Journal of Cellular Biochemistry, 120(2), 1943-1957.


- . Journal of Genetic Engineering and Biotechnology, 16(2), 427-432.


- . International Journal of Nanomedicine, 13, 8137-8151.


- . EBioMedicine, 35, 106-113.


- . Redox Biology, 18, 114-123.


- . Trends in Genetics, 34(8), 574-577.


- . Free Radical Biology and Medicine, 126, 153-165.


- . Oncotarget, 9(53), 30053-30065.


- . Cell Reports, 23(11), 3352-3365.


- . Journal of Cellular Biochemistry, 119(8), 6869-6881.


- . Brain, 141(5), 1247-1262.


- . Nature Reviews Molecular Cell Biology, 19(6), 343-344.


- , 173-181.


- . Neuron, 96(4), 730-735.


- . Nature Neuroscience, 20, 1225-1235.


- . Disease Models & Mechanisms, 10, 859-868.


- . Nature Communications, 8.


- . Science Advances, 3(4).


- . Nucleic Acids Research, 45(3), 1159-1176.


- , 41-58.


- . Biosensors and Bioelectronics.


- . Mechanisms of Ageing and Development.


- . Scientific Reports, 6, 26626-26626.


- . Cell Reports, 15(4), 893-908.


- . Oncoscience, 2(3), 294-308.


- . Nature Reviews Cancer, 15(3), 137-151.


- . Human Molecular Genetics, 24(3), 828-840.


- , 39-74.


- . Molecular Cancer Therapeutics, 14(2), 575-585.


- . Nature Genetics, 46(5), 516-521.


- . Analytical Biochemistry, 454, 17-22.


- . Nucleic Acids Research, 42(5), 3089-3103.


- . PLoS One, 8(4), e58239.


- . Nucleic Acids Res, 40(17), 8371-8380.


- . Biochimie, 94(8), 1749-1753.


- . Nat Commun, 3, 733.


- . Br J Cancer, 106(1), 18-24.


- . Curr Med Chem, 19(23), 3874-3885.


- . EMBO Mol Med, 3(2), 78-88.


- . J Biol Chem, 286(1), 403-409.


- . Hum Mol Genet, 19(7), 1324-1334.


- . Cell Cycle, 9(3), 588-595.


- . Nature, 461(7264), 674-678.


- . Nat Neurosci, 12(8), 973-980.


- . DNA Repair (Amst), 8(6), 760-766.


- . Biochem Soc Trans, 37(Pt 3), 577-581.


- . Mol Cell Biol, 29(5), 1354-1362.


- REPAIR OF DNA SINGLE-STRAND BREAKS IN NEURONS: IMPLICATIONS ON HUMAN HEALTH. CELLULAR ONCOLOGY, 31(2), 131-131.


- . Molecular and Cellular Biology, 28(10), 3561.


- . EMBO J, 26(22), 4720-4731.


- Characterisation of aprataxin- and PNK-like factor: a novel FHA-domain protein involved in single and double-strand break repair. MUTAGENESIS, 22(6), 449-450.


- . DNA Repair (Amst), 6(10), 1485-1495.


- . Mol Cell Biol, 27(10), 3793-3803.


- . Neuroscience, 145(4), 1260-1266.


- . Nature, 443(7112), 713-716.


- . Mutagenesis, 21(4), 219-224.


- . Methods Enzymol, 409, 410-425.


- . NATURE, 434(7029), 108-113.


- . Cell, 117(1), 17-28.


- . Nucleic Acids Res, 31(19), 5526-5533.


- . Nucleic Acids Research.


- . BMC Medical Genomics, 17(1).


- . Advanced Science.


- . Scientific Reports, 14(1), 11840.


- . Physical & Occupational Therapy In Geriatrics, 1-24.


- . Biochemical Journal.


- . Journal of Nanosciences: Current Research, 01(01).


- . Journal of Cancer Science & Therapy, 06(07).


- . Cancers, 13(5), 944-944.


- . Cellular and Molecular Life Sciences.


- . Journal of Clinical Pathology.


Chapters
- , Paraproteinemia and Related Disorders (pp. 57-77). Springer International Publishing


- Gene Therapy in the Nervous System: Failures and Successes In El-Khamisy S (Ed.), Personalised Medicine Lessons from Neurodegeneration to Cancer Springer


- In El-Khamisy (Ed.), Personalised Medicine


- Personalised Medicine Lessons from Neurodegeneration to Cancer Preface, PERSONALISED MEDICINE: LESSONS FROM NEURODEGENERATION TO CANCER (pp. V-V).


- , Advances in Experimental Medicine and Biology (pp. 157-178). Springer International Publishing


- , Movement Disorders (pp. 1033-1041). Elsevier


- , Movement Disorders: Genetics and Models: Second Edition (pp. 1033-1041).


Conference proceedings papers
- Immunohistochemical analysis of R-loop burden in a cohort of Oropharyngeal Squamous Cell Carcinomas (OPSCC) with a poor prognosis. VIRCHOWS ARCHIV, Vol. 485 (pp S404-S405)


- The Role of R-loops in the Development of Cisplatin Resistance in Oropharyngeal Squamous Cell Carcinoma (OPSCC). JOURNAL OF PATHOLOGY, Vol. 261(SUPPL1) (pp S62-S62)


- Exploration of the transcriptomic landscape of HPV-positive and HPV-negative oropharyngeal squamous cell carcinoma upon development of cisplatin resistance. VIRCHOWS ARCHIV, Vol. 481(SUPPL 1) (pp S73-S73)


- Modulating the polymorphic UGT1A1 gene to enhance camptothecin anti-colorectal effect. CANCER RESEARCH, Vol. 82(12)


- . Molecular and Cellular Biology/Genetics


- BRCA1 and BRCA2 mutations and its clinical relevance among egyptian breast cancer women.. CANCER RESEARCH, Vol. 81(13)


- The effect of E6 and E7 knockdown and PARP inhibition on cisplatin sensitivity in oropharyngeal squamous cell carcinoma. VIRCHOWS ARCHIV, Vol. 477 (pp S76-S76)


- . ACM International Conference Proceeding Series (pp 391-396)


- DNA Damage and Repair in Patients with Cryoglobulinemic Vasculitis Treated with Direct Anti-HCV Drugs. ARTHRITIS & RHEUMATOLOGY, Vol. 70


- COMPARATIVE MIRNA MICROARRAY PROFILING INDICATES MIR-363 PROMOTES CHEMORESISTANCE IN OVARIAN CANCER CELLS BY TARGETING THE HIPPO MEMBER, LATS2. INTERNATIONAL JOURNAL OF GYNECOLOGICAL CANCER, Vol. 27 (pp 1952-1952)


- . Molecular and Cellular Biology, Genetics


- Treatment of Cryoglobulinemic Vasculitis with Sofosbuvir in Four Combination Protocols. ARTHRITIS & RHEUMATOLOGY, Vol. 68


- Tyrosyl-DNA phosphodiesterase 1-a new player in base excision repair. FEBS JOURNAL, Vol. 280 (pp 63-64)


Preprints
- . Life Science Alliance, 5(8), e202101145-e202101145.
- Research group
-
I welcome applications from self-funded prospective home and international PhD students; see examples of possible projects below.
You can apply for a PhD position in MBB here.
Contact me at S.El-Khamisy@sheffield.ac.uk for further information.
- Teaching activities
-
Level 3 Modules
MBB313 Genome Stability and Genetic Change
Level 2 Modules
MBB262 Genetics 2